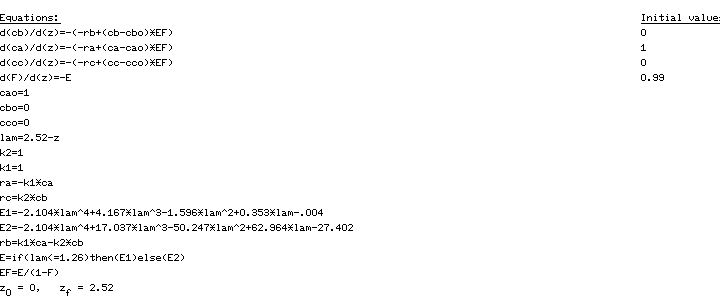occurring in two different reactors with the same mean residence time tm=1.26 min, but with the following residence-time distributions which are quite different:
(a) Calculate the conversion predicted by an ideal(b) Fit a polynomial to each RTD (Figures CDE13-1.1 and CDE13-1.2). Note: These curves are identical to Figures E13-9.1 and E13-9.2 in the text.
- PFR.
- CSTR.

Figure CDE13-1.1
E1 (t ):asymmetric distribution.

Figure CDE13-1.2
E2 (t ): bimodal distribution.
(c) For the multiple reaction sequence given above, determine the product distribution in each reactor for 1. The segregation model. 2. The maximum mixedness model.
Solution
PFRCombining the mole balance and rate laws for a PFR reactor, we have

(CDE13-1.1)
(CDE13-1.2)
(CDE13-1.3)
The initial conditions are V = 0, CA = 1, CB = CC = 0. In order to compare the performance of the different models and different RTDs, the mean residence time, , was set equal to 1.26 min.
, was set equal to 1.26 min.
The POLYMATH program used to solve this PFR system is shown in Table CDE13-1.1. The solution is CA = 0.284, CB = 0.357, CC = 0.359, X = 71.6%.