|
What are the relative contributions of cis- and trans-regulatory changes to expression differences genome-wide and how does natural selection shape their evolution?
We are determining the extent and inheritance of expression differences over a range of divergence times, assessing the relative contributions of cis- and trans-regulatory changes to differential expression, and identifying genomic features that correlate with particular types of regulatory changes. We are also investigating the roles of natural selection and genetic drift in creating extant patterns of regulatory variation.
Next-generation sequencing is a powerful tool with which to address these issues. By extracting RNA, converting it into cDNA, and performing deep sequencing of these cDNA libraries - a technique known as RNA-seq - we can infer the relative abundance of transcripts from individual genes as well as individual alleles (as long as they differ by at least one transcribed base) (Fontanillas et al. 2010; McManus et al. 2010; Coolon et al. 2012). Comparing differences in total gene expression between two inbred genotypes to differences in allele-specific expression in F1 hybrids (produced by crossing these two inbred genotypes) allows the relative contributions of cis- and trans-acting changes to beinferred (Wittkopp et al. 2004; 2008). Comparing total expression levels between two genotypes and their F1 hybrids assesses the dominance of regulatory alleles (Landry et al. 2005; McManus et al. 2010). Combining these measurements with genomic sequence data can identify correlated factors that might predict genetic mechanisms underlying expression differences for individual genes. Such information can be used in a medical context to study expression differences that cause disease states.
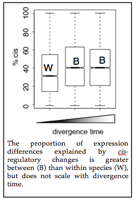
One of the most intriguing patterns to emerge thus far from my studies of regulatory evolution is that cis-regulatory changes explain a greater proportion of expression differences between than within Drosophila species (see Figure) (Wittkopp et al. 2008); this same pattern was recently seen in yeast (Emerson et al. 2010), suggesting its generality. To determine whether this difference (and other aspects of gene regulation) increase systematically with divergence time or change qualitatively with speciation, we are comparing gene expression in strains and species of Drosophila with divergence times ranging from 10,000 to 5,000,000 years ago (Coolon et al. in prep). We have also used these data to show an absence of imprinting in D. melanogaster (Coolon et al. 2012).
Currently, we are using these data to (1) identify properties of genomic sequence that correlate with specific types of regulatory changes and (2) select genes with lineage-specific changes in cis-regulation suitable for finding the specific nucleotides responsible for functional divergence.
| 




