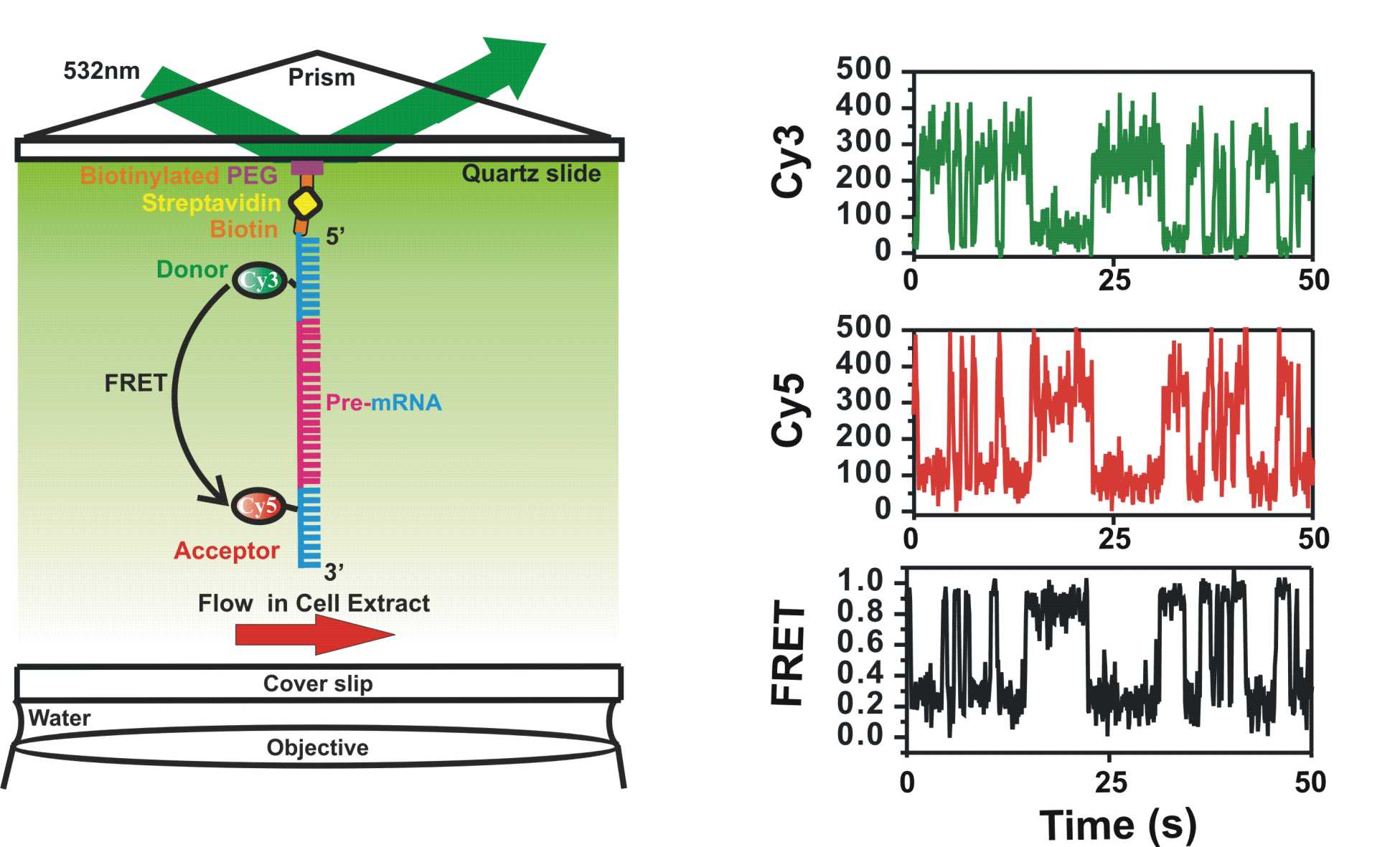
Eukaryotic pre-messenger RNA (pre-mRNA) undergoes several maturation steps en route to the ribosome for translation. A particularly important step in this pathway is splicing, where the intervening sequences (introns) are removed from the pre-mRNA and the coding sequences (exons) are ligated together. This process involves two transesterification reactions catalyzed by a multimegadalton RNA- protein complex termed the Spliceosome. The spliceosome is a large ribonucleoprotein complex (RNPs) which contains a total of over 100 different proteins, and 5 small nuclear RNAs (snRNAs). Biochemical and genetic characterization of pre-mRNA splicing has revealed that conformational rearrangements within the spliceosome as well as changes in its composition are required throughout the assembly and catalytic processes. Although much attention has been placed on the spliceosome itself, there is a lack of understanding regarding the positioning and conformational dynamics of the pre-mRNA throughout the splicing process. Using single-molecule Fluorescence Resonance Energy Transfer (sm-FRET), we have been able to observe real-time dynamics of spliceosome assembly in a manner that previously was not possible. By fluorescently labeling the pre-mRNA substrate, spliceosomal proteins and snRNAs, we will for the first time learn how the pre-mRNA’s conformation is related to the different steps in catalysis and spliceosomal assembly.