|
|
|
How do cis-regulatory sequences determine gene expression and how does the architecture of cis-regulatory regions change over time?
When, where, and how much a gene is expressed is controlled primarily by the interaction of cis-regulatory DNA sequences with trans-regulatory transcription factors (TFs). Changes in TF binding sites contribute to disease states, phenotypic diversity among healthy individuals, and divergent phenotypes between species. Despite their importance, many questions remain about the way in which cis-regulatory DNA sequences translate TF binding into transcriptional activity (Wittkopp and Kalay 2012). The rapid evolution of yellow enhancers provides us with an excellent opportunity to study the evolution of cis-regulatory sequences (Wittkopp et al. 2002; Gompel et al. 2005).
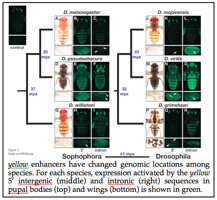
Using reporter genes in D. melanogaster, we discovered that the locations of tissue specific enhancers of the pigmentation gene yellow have changed positions in the genome among species (see Figure) (Kalay and Wittkopp 2010). Sequence analysis suggested that this was more likely due to the gradual gain and loss of transcription factor binding sites than duplications or rearrangements. This allows us to use these regions to compare DNA sequence, TF binding, and cis-regulatory activity among sequences that have independently acquired the same function as well as among orthologous DNA sequences whose functions have diverged.
Currently, we are (1) analyzing yeast-1-hybrid data identifying TFs that bind to species-specific enhancers of yellow, (2) localizing sequences responsible for tissue-specific activity within each enhancer region, and (3) using information gained from this work to elucidate developmental mechanisms controlling abdominal patterning during pupal development.
|





