



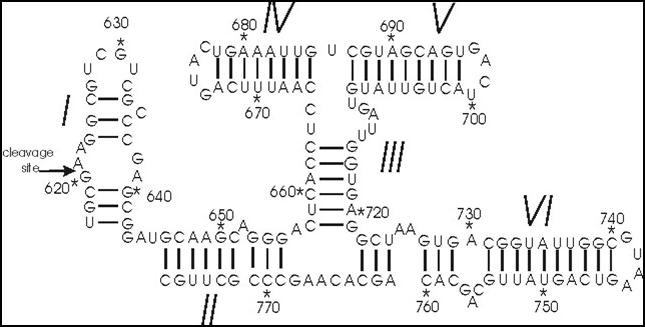
The Varkud satellite (VS) ribozyme is a naturally occurring RNA molecule, found in the bread mold Neurospora, that catalyzes the self-cleavage and ligation reactions necessary for the life cycle of the VS RNA. It is the largest known member of the small self-cleaving ribozymes and the only one in this class for which the structure is not known at atomic resolution. Using single-molecule fluorescence resonance energy transfer (smFRET), we have studied how the molecule’s global dynamics influence its function as a catalyst1. However, to gain more detailed insights into the VS ribozyme’s three-dimensional structure, dynamics, and catalytic mechanism, a high-resolution X-ray crystal structure of the complete molecule is crucial. To this end, we are trying to crystallize the VS ribozyme using a novel method of purifying the ribozyme in its native state without subjecting it to denaturing conditions. By avoiding the harsh conditions of traditional purification by denaturing gel electrophoresis, which can cause the molecule to refold into erroneous structures, we hope to isolate a conformationally homogeneous population more conducive to crystallization and structural studies.
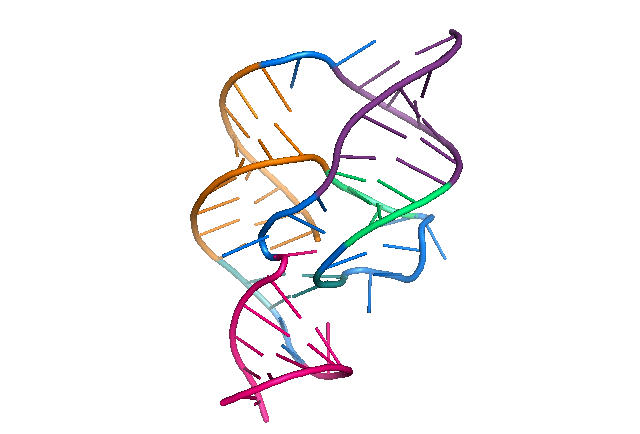
|
The hepatitis delta virus (HDV) ribozyme is the only small self-cleaving ribozyme known to exist within a human pathogen. The activity of two ribozyme variants, the genomic and antigenomic HDV ribozymes, is essential for replication of the HDV, which can lead to liver cirrhosis and death. Furthermore, structurally similar RNA motifs have been reported in the genomes of a wide variety of organisms, including that of humans1, so understanding the folding and function of the HDV ribozyme has broad implications for biology and medicine. Previously, we have shown that an HDV ribozyme undergoes a conformational change that accompanies catalysis in solution2,3, discovered a common U-turn motif at the active site that appears closely connected to this conformational switch4, and examined the possible role of an active-site cytosine residue as the catalytic general base5. Using a combination of single-molecule FRET and molecular dynamics, we are now seeking a deeper understanding of the dynamics underlying catalysis in the HDV and related ribozymes over time scales ranging from picoseconds to minutes. |
1. Webb, C. T., Riccitelli, N. J., Ruminski, D. J., Lupták, A. (2009). Widespread occurrence of self-cleaving ribozymes. Science 326, p. 953.
2. Pereira, M.J.B., Harris, D.A., Rueda, D., and Walter, N.G. (2002) Reaction pathway of the trans-acting hepatitis delta virus ribozyme: A conformational change accompanies catalysis. Biochemistry 41, p. 730-740.
3. Harris, D.A., Tinsley, R.A., and Walter, N.G. (2004) Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 341, p. 389-403.
4. Sefcikova, J, Krasovska, M.V., Sponer, J., and Walter, N.G. (2007) The genomic HDV ribozyme utilizes a previously unnoticed U-turn motif to accomplish fast site-specific catalysis. Nucleic Acids Res. 35, p. 1933-1946.
5. Banas, P., Rulisek, L., Hanosova, V., Svozil, D., Walter, N.G., Sponer, J., and Otyepka, M. (2008) General base catalysis for cleavage by the active-site cytosine of the hepatitis delta virus ribozyme: QM/MM calculations establish chemical feasibility. J. Phys. Chem. B 112, p. 11177-11187.