Antifouling Polymers
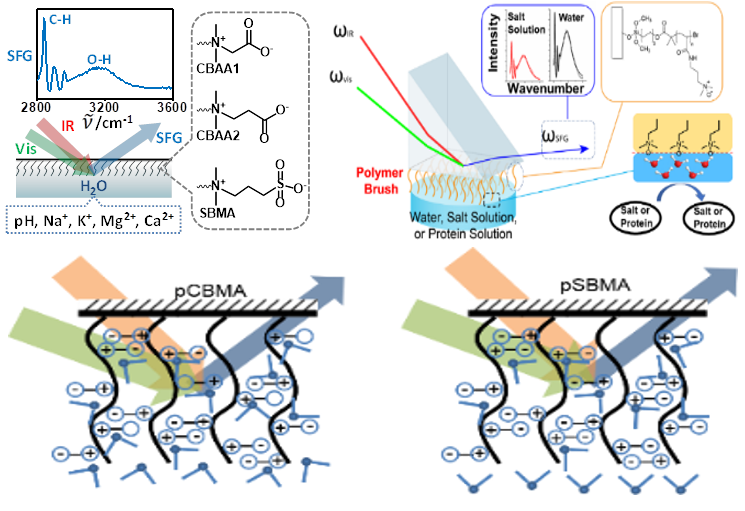
Antifouling polymers play important roles in a variety of applications such as biomedical implants, drug delivery, membranes for separation, and coatings used in the marine environment, etc. Recently, zwitterionic polymers have been developed into powerful environmentally friendly marine antifouling materials with many advantages to replace current toxic coatings.
It is widely believed that the excellent antifouling performance of zwitterionic polymers is due to the strong surface hydration, which prevents other molecules from displacing surface bound water to stick to the surface.
We are using SFG to study the relationship between surface hydration and state-of-the-art antifouling zwitterionic polymer coatings. The effects of the distance between the positive and negative charges on the interfacial hydration and the antifouling performance of zwitterionic polymers are under the current investigation. We also aim to understand the impact of charge delocalization on the zwitterionic polymer antifouling activity.
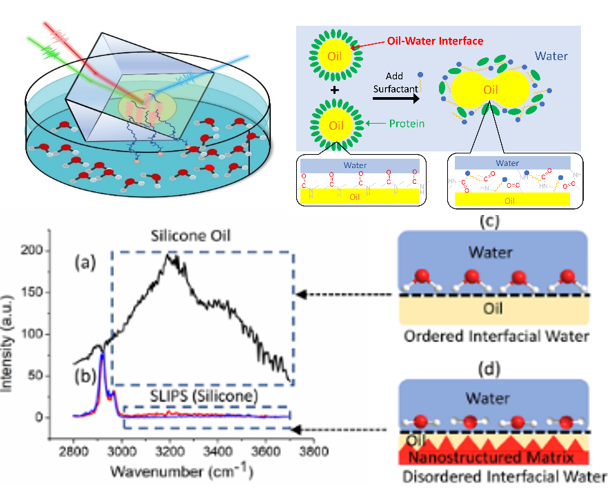
In addition to the zwitterionic polymers, we are studying various amphiphilic polymers, nanostructured coatings, and silicone materials for antifouling and fouling-release purposes.